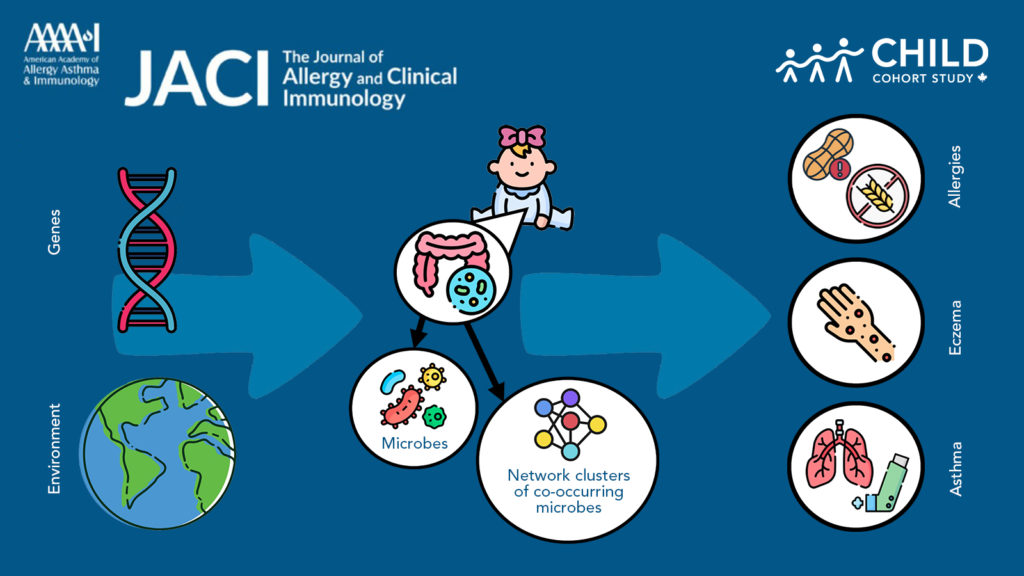
New research from CHILD and the Computational Genomics Laboratory at Queen’s University sheds light on how interactions between our genes and early-life environment impact our gut microbes and risk of asthma and allergies.
Published in The Journal of Allergy and Clinical Immunology (JACI), these new findings highlight the importance of considering human genomics in microbiome research—the study of how the microbes living in and on our bodies influence our health. This new research also identifies several early-life exposures that influence allergy and asthma risk, informing on potential early-life risk factors, which could help in the development of preventative interventions.
“Our study supports past research findings from CHILD and other studies revealing important associations between gut microbes and risk of developing asthma or allergies,” comments first author Sara Stickley, a PhD candidate in the Experimental Medicine program of the Department of Biomedical and Molecular Sciences at Queen’s University.
“Earlier CHILD research also demonstrated how environmental factors—like whether babies are born vaginally or by c-section, antibiotics use, or whether there are furry pets in the home—impact our gut microbiome.”
“Our new study is among the first to look at early-life influences on the infant gut microbiome through a genomics lens: adding the crucial consideration of our genetic predispositions to the puzzle.”
For this study, researchers analyzed the bacterial content of stool samples taken from nearly 800 CHILD participants at 3 months and 1 year of age. Using a machine-learning algorithm, they identified clusters of correlated microbes in the stool associated with the development of asthma or allergic sensitivity by age 5 years. Participants were assessed for indications of asthma and allergic sensitivity at age 5 years through medical records as well as tests conducted by CHILD clinicians.
The researchers next performed genome-wide association studies (GWAS) to identify genomic variations associated with gut microbiota among the infants—focusing on those bacteria associated with asthma and allergy risk.
Then they analyzed how various environmental exposures during childhood potentially interact with genomic variations to influence gut microbiota of the infants. To do this, they used information CHILD had collected about environmental exposures from hospital records, questionnaires completed by the parents of participants when they were babies, and comprehensive assessments conducted in each family’s home.
“We were able to clearly identify gut microbes associated with respiratory and allergy-related outcomes, some of which differed between girls and boys,” states lead researcher Dr. Qingling Duan, a Queen’s National Scholar in Bioinformatics and head of the Computational Genomics Laboratory at Queen’s University. “And we were able to further link these microbes with the participants’ genomics and early-life environmental exposures.”
For example, they found that the gut bacterium known as Blautia obeum was associated with variants in the MARCO gene and a lower prevalence of food sensitizations among the children. They also found variants in the SMAD2 gene were associated with a cluster of gut bacteria implicated with asthma risk; however, this genetic association varied with breastfeeding duration among the infants.
These findings improve our understanding of the mechanisms by which genomic and environmental factors contribute to risk of asthma and atopic sensitizations, which could inform new strategies for prevention and treatment of such diseases in the future, the researchers note.
“It is a major challenge for health researchers to parse out all the genomic and non-genomic risk factors, their relative importance, and their interactions with each other,” adds Dr. Duan.
“We are tremendously fortunate to have a research platform like CHILD available to us, and to have families as generous as those involved in CHILD. Few studies have collected so much data over many years from such a large sampling of people, allowing us to conduct such innovative and nuanced studies of factors contributing to complex diseases.”
